Physics of Living Matter
We use experiments, computer simulations, and theoretical calculations to understand how physical laws affect living organisms.
Overview
Our research in this area spans many length and time scales: from aqueous solutions of small bioactive molecules through proteins and DNA to single cells, cell-cell interactions, and collection of organisms in ecosystems, studying phenomena occurring at picoseconds to millions of years. Many experiments use optical techniques made available through COSMIC. In our wet labs (biological hazard containment level 1 and 2) we routinely work with bacteria such as E. coli, B. subtilis, and P. aeruginosa, as well as extremophile microbes that can survive at extremes of temperature, or pressure and can even eat rocks! We also use microfluidics and plate readers for high-throughput experiments, and neutron scattering for subcellular-resolution experiments.
Among the powerful resources accessed by our computer simulators and theorists is the computer cluster Eddie hosted by The Edinburgh Compute and Data Facility ECDF as well as the supercomputers ARCHER2 and Cirrus hosted by the Edinburgh Parallel Computing Centre (EPCC).
Much of the research is closely tied to our programme on soft matter and statistical physics and complexity. Funding comes from EPSRC, BBSRC, BEIS, Scottish Enterprise, Wellcome Trust, The Royal Society, and the Royal Society of Edinburgh.
Physics of Living Matter forms part of the Scottish Universities Physics Alliance (SUPA) Physics and Life Sciences (PaLS) theme. The SUPA Graduate School offers annual Prize Studentships for PhD study (the typical deadline is end of January for entry in September).
Research interests
Protein self-assembly
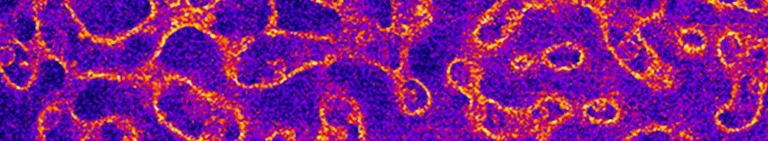
All life is made of proteins - long chains of polymerized aminoacids which fold into complicated 3d structures with various shapes and functions. In order to function properly, a protein must fold into an appropriate configuration, otherwise it will not function properly and may result ina disease. Alzheimer's disease, Parkinson's disease are just two examples of what happens when a protein folds incorrectly. We use experiments and molecular dynamics simulations to investigate this self-assembly process of proteins. We are particularly interested in how this process can be affected by environmental factors. Besides proteins implicated in human diseases we investigate bacterial proteins which enable microbes to form "coats" protecting them from external stresses such as antibiotics.
DNA folding and transcription regulation
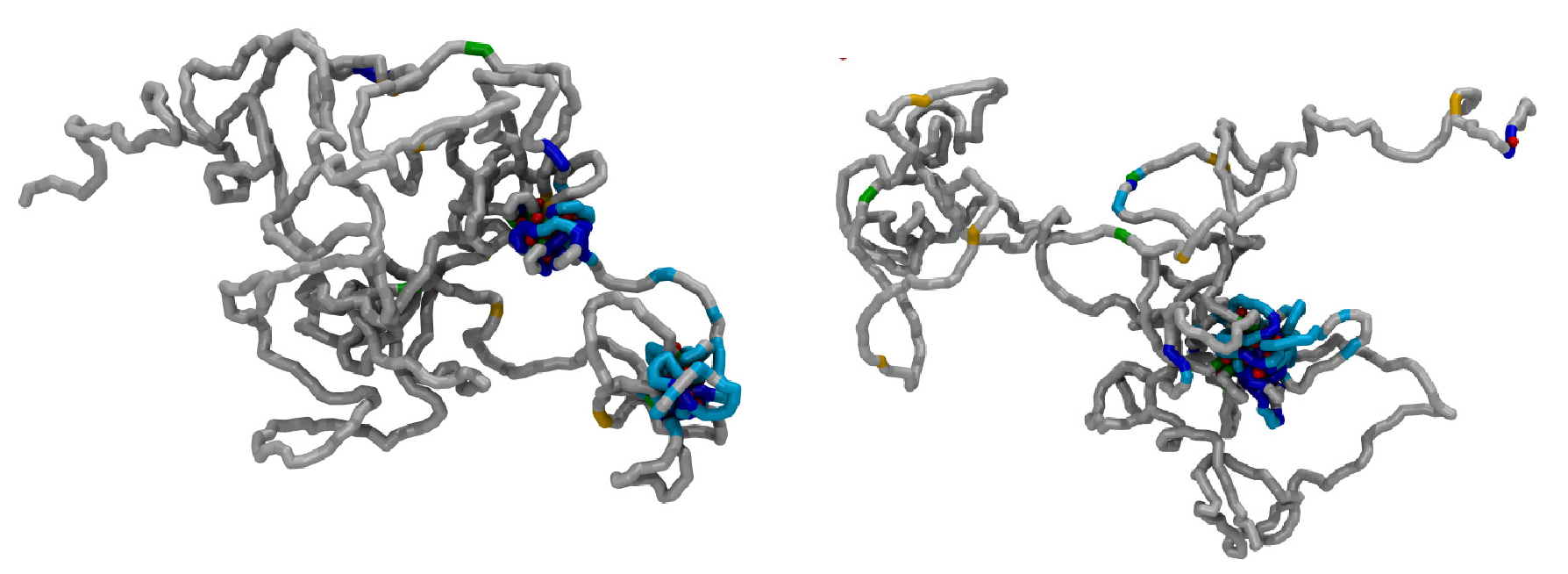
DNA is arguably one of the most important biopolymers, as its sequence encodes the genetic instructions needed in the development and functioning of living organisms. Within human nuclei, DNA is organised into chromatin, a fiber made up by histone proteins around which the DNA wraps essentially due to electrostatic interactions. Little is known either about the higher order structure of chromatin in the cell, or even how it self-assembles dynamically within the nucleus or even in the test tube.
Within ICMCS, we model DNA, chromatin and chromosomes at different levels of detail. Current projects involve modelling at different levels of resolution of chromosomes, or fractions thereof, in yeast, Drosophila (the fruit fly), and human. Finally, we plan to study the dynamics of such structures, and how the structures change when the genes are transcriptionally active or inactive.
Protein films & shells
Viruses have unique mechanical properties: they have to be strong enough to withstand the stresses involved in reaching their target in a host but fragile enough that disassembly and release of genetic material can be triggered by physicochemical stimulli on reaching the target. We are investigating the physical principles behind these mechanical properties using a combination of neutron spin echo spectroscopy. We also use our armoury of neutron & x-ray techniques to investigate the assembly of protein & peptide films at fluid interfaces.
Cell membranes

Bacterial resistance to antibiotics has in 2013 been placed on the National Risk Register and we use neutron & x-ray reflectivity to identify unifying physical principles in the way in which peptides involved in controlling bacterial growth and cell division interact with bacterial membranes and how this is modulated by the cell's metabolic state via membrane potential. The molecular-scale experimental information is nicely complemented by coarse-grained molecular dynamics simulations.
Cellular motility
Many cells are motile: unicellular organisms move around searching for food or evading toxic chemicals and predators, whereas eukaryotic (animal) cells such as the sperm or macrophages (white blood cells) search for the egg or invading microbes. We are interested how cytoskeleton dynamics (animal cells) or the rotation of the flagella (bacteria) is translated into the motion of the cell. This process involves physical interactions (hydrodynamics, adhesion) between the cell and its environment. We therefore investigate bacterial and sperm motility in different media using differential dynamic microscopy (DDM). We also develop theoretical and computer models of cellular motility.
We also use motile bacteria and algae as model 'active colloids' to study the physics of systems of self propelled particles. Current statistical mechanics offers no general recipe for determining the collective behaviour of such 'active matter', and experiments using well defined suspensions of motile microorganisms constitute a good source of data to guide theory development.
Bacterial colonies and biofilms
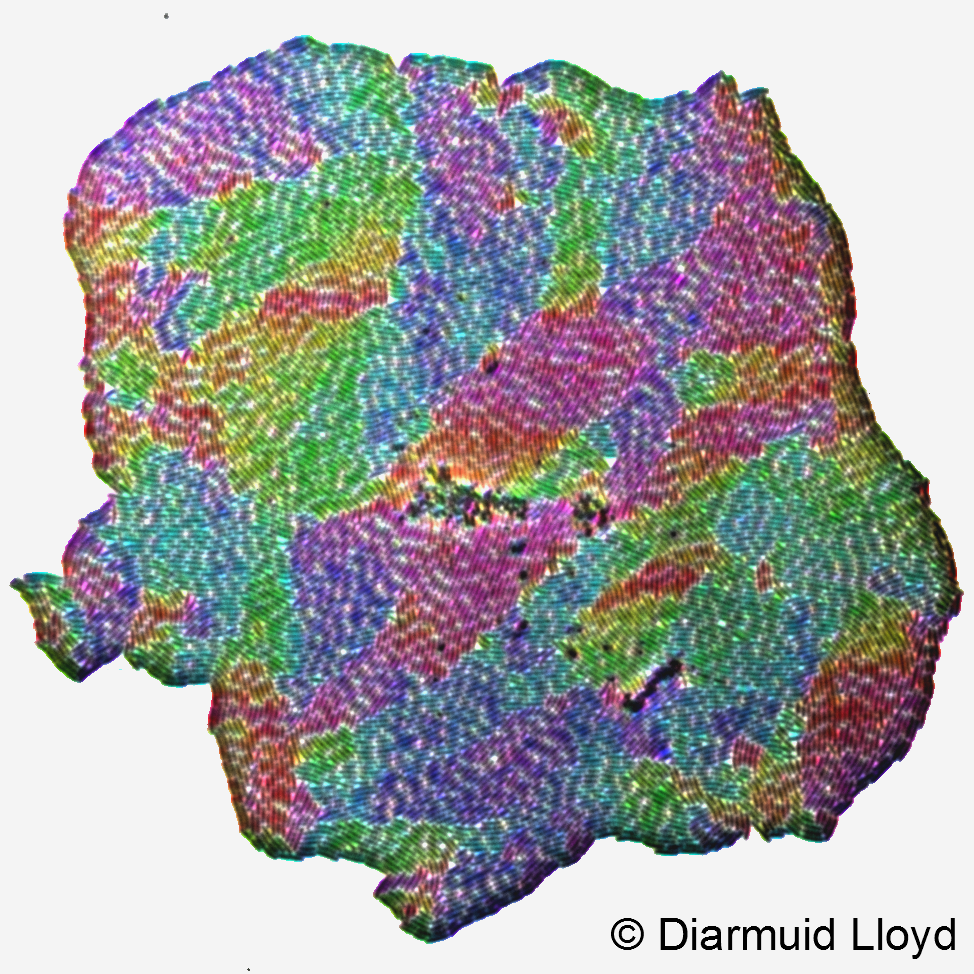
Bacteria often attach to surfaces and form colonies and biofilms. Surface-growing microbes are important in medicine (tooth plaque, infected catheters or implants) and industry (growth on food or in water distribution systems). Colonies of rod-shaped bacteria can also be viewed as the active nematics - an interesting counterpart to nematic liquid crystals. We are interested in how physical properties of bacterial cells and their environment affect how such colonies grow. We use experiments (microscopy) and computer simulations to address a variety of questions: what determined the shape of colonies and the orientation of cells in a growing colony, how colonies form multiple layers of cells, and how these processes affect the fate of new mutant bacteria occurring spontaneously in the colony. We also use agent-based simulations to predict the structure of bacterial biofilms under a variety of environmental conditions.
Microbial growth and evolution
Microbes (micro-size unicellular organisms such as bacteria) are able to grow in a wide range of conditions and utilize many different nutrients. Microbes can also grow in the presence of harmful chemicals such as antibiotics. We are interested in how physics and chemistry limit these capabilities. In particular, we investigate how bacteria respond and evolve resistance to antibiotics. In our research we use a combination of macro- and microfluidics experiments, computer simulations, and analytical calculations.
Life in Extreme Environments
All environments subject organisms to various combinations of physical and chemical stressors. We are interested in how life adapts to extreme conditions, for example in the deep subsurface or in newly available habitats, and how it copes with multiple extremes and energy limitation in such places. This work is primarily laboratory based as well as using environmental microbiology and modelling. Our work also involves studying what determines microbial community structure in extreme environments. This work has applications to understanding the limits of life on Earth and assessing the habitability of extreme environments on Earth and potentially elsewhere. For more details see UK Centre for Astrobiology.
Statistical-physics models of living systems
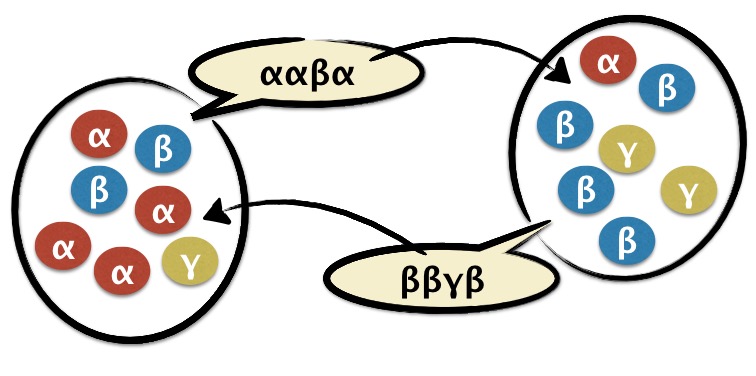
We use techniques borrowed from equlibrium and non-equilibrium statistical mechanics to study how complex systems of living organisms evolve in time and space. Among the systems we study are simple models of cultural evolution, the evolution of language, Darwinian evolution of spatially heterogeneous populations. See Statistical Physics and Complexity for more details.
Techniques
Below is a list of some of the techniques we use in our research:
- Optical microscopy: brightfield, fluorescence, confocal
- Neutron scattering and reflectivity
- Molecular/Brownian dynamics/agent-based simulations
- Mathematical modelling: ordinary and partial differential equations, stochastic models
PhD project opportunities in Physics of Living Matter
- Active-Nematic/Polymeric Composites
- Bacteria Motility in Complex Media
- Bacterial Biofilm Matrix Remodelling in Complex and Dynamic Environments
- Communal Living for Bacteria
- Controlling bacteria aggregation for environmental remediation and biotechnological applications
- Controlling space and time in active matter
- Controlling the microbial generation of nitrous oxide in wastewater treatment; a pathway to reduce greenhouse gas emissions
- Deciphering species coexistence using microbial microcosms
- Developing tractable model systems for filamentous bacteria in wastewater treatment
- Enhancing cardiovascular long axial field-of-view positron emission tomography using photon-counting computed tomography
- Evolution of Life in Droplets
- Force microscopy and spectroscopy of chromatin assemblies in disease
- How antibiotics kill persistent infections
- Microswimmers in Complex Fluids
- Modelling the impact of extreme events on the emergence of infectious disease variants
- Moving and growing among entanglements: the role of DNA in the physics of bacterial swimming and aggregation
- Physics of genome structure in health and disease
- Shaping the genome through formation of protein droplets
- Statistical physics of dividing and differentiating stem cell populations
- Topologically Active Polymers (Experimental and/or Computational)
- Using Viruses to Kill Bacteria
People in Physics of Living Matter
Telephone numbers in the list below are shown as UK numbers. Callers from outside the UK should remove the leading zero and use the UK country code (+44).
| Name | Position | Contact details | Location | Photo |
|---|---|---|---|---|
| Academic staff | ||||
| Graeme Ackland | Professor | gjackland [at] ed.ac.uk | JCMB 2602 |  |
| Richard Blythe | Professor | R.A.Blythe [at] ed.ac.uk | JCMB 2505 |  |
| Chris Brackley | Lecturer in Theoretical/Computational Condensed Matter Physics | C.Brackley [at] ed.ac.uk | JCMB 2612 |  |
| Aidan Brown | Lecturer | Aidan.Brown [at] ed.ac.uk | JCMB 2616 |  |
| Paul Clegg | Professor | paul.clegg [at] ed.ac.uk | JCMB 2614 |  |
| Charles Cockell | Professor of Astrobiology | C.S.Cockell [at] ed.ac.uk | JCMB 1503 |  |
| Jean-Christophe Denis | Lecturer & Public Engagement Officer | J.C.Denis [at] ed.ac.uk | JCMB 2905 |  |
| Martin Evans | Professor of Statistical Physics | m.evans [at] ed.ac.uk | JCMB 1510 |  |
| Cait MacPhee | Professor | cait.macphee [at] ed.ac.uk | JCMB 2613 |  |
| Davide Marenduzzo | Personal Chair in Computational Biophysics | Davide.Marenduzzo [at] ed.ac.uk | JCMB 2506 |  |
| Gavin Melaugh | Chancellors Fellow | G.Melaugh [at] ed.ac.uk | JCMB 1506 | |
| Alexander Morozov | Professor | Alexander.Morozov [at] ed.ac.uk | JCMB 2507 |  |
| Wilson Poon | Emeritus Professor | W.Poon [at] ed.ac.uk | JCMB 2617 |  |
| Tyler Shendruk | Reader in Theoretical/Computational Condensed Matter Physics | t.shendruk [at] ed.ac.uk | JCMB 2422 | |
| Simon Titmuss | Lecturer | Simon.Titmuss [at] ed.ac.uk | JCMB 2504 |  |
| Willem Vanderlinden | Reader | willem.vanderlinden [at] ed.ac.uk | JCMB 1612 | |
| Research staff | ||||
| Jochen Arlt | Senior Lecturer | j.arlt [at] ed.ac.uk | JCMB 2601 |  |
| Davide Michieletto | Royal Society Research Fellow | Davide.Michieletto [at] ed.ac.uk | JCMB 1505 |  |
| Ryan Morris | PDRA | ryan.morris [at] ed.ac.uk | JCMB 1601 | |
| Giuseppe Negro | Lecturer in Theoretical/Computational Biophysics | giuseppe.negro [at] ed.ac.uk | JCMB 1611 | |
| Jana Schwarz-Linek | Senior Researcher | Jana.Schwarz-Linek [at] ed.ac.uk | JCMB 2609 |  |
| Tiffany Wood | Senior Knowledge Transfer Fellow | tiffany.wood [at] ed.ac.uk | JCMB 2611 |  |
| Research postgraduates | ||||
| Joseph M. Knight | Postgraduate Student | Joseph.Knight [at] ed.ac.uk | JCMB 2510 | |
| Auro Varat Patnaik | Postgraduate Student | Auro.Patnaik [at] ed.ac.uk | JCMB 1511 | |
| Anne-Maelle Penot | Postgraduate Student | A.M.M.Penot [at] sms.ed.ac.uk | JCMB 1511 | |