PhD project: Topologically Active Polymers (Experimental and/or Computational)
Project description
General aim of the projects:
Inspired by how DNA is organised inside our cells, in this project the student will investigate new types of soft materials and complex fluids that are made by polymers which change topology in time. While most of the polymer-based materials that we use every day such as creams, gels and plastics are made by polymers which do not change their architecture or topology (or do so randomly), the DNA inside our cells is precisely mechanically and topologically manipulated by sophisticated proteins. The aim is to design and study systems of polymers which can undergo topological operations that are inspired by those performed by proteins in the cells. The grand goal of this investigation is to create new "topologically active" soft materials which may be patented or find applications in bio- and nano-technology such as drug delivery or cellular scaffolds.
Experimental Project: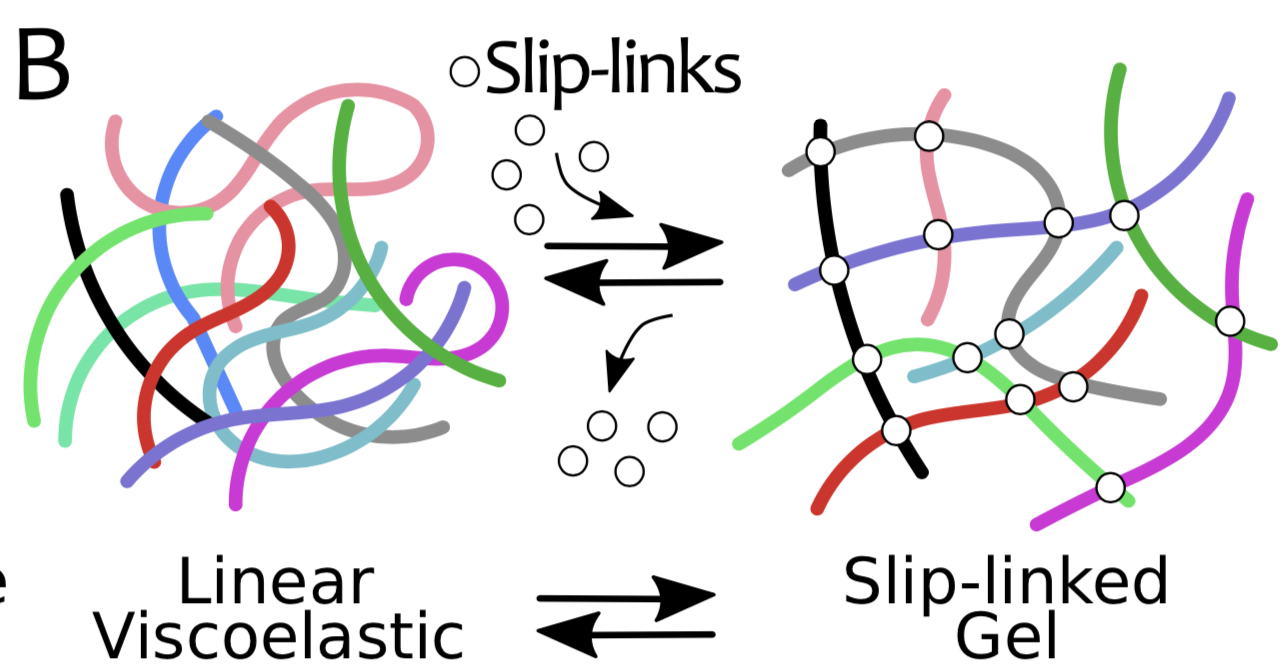
The student will prepare entangled systems of DNA either purchased from companies or extracted from bacteria in a variety of topological conditions, i.e. linear, circular, supercoiled, knotted or linked. S/he will then study the rheological properties of such a complex fluid once certain classes of proteins (such as Topoisomerase, restriction enzymes, ligases, or others) are added into the system. S/he will be mostly performing microrheology -- a technique based on tracking the diffusion of beads embedded in the fluid using microscopy -- and (confocal) imaging of fluorescent DNA. Variations of this project may involve the design and investigation of the rheological properties of entangled solutions of DNA origami and are connected with the computational porject on "chimeric" polymers (see below).
Computational Project:
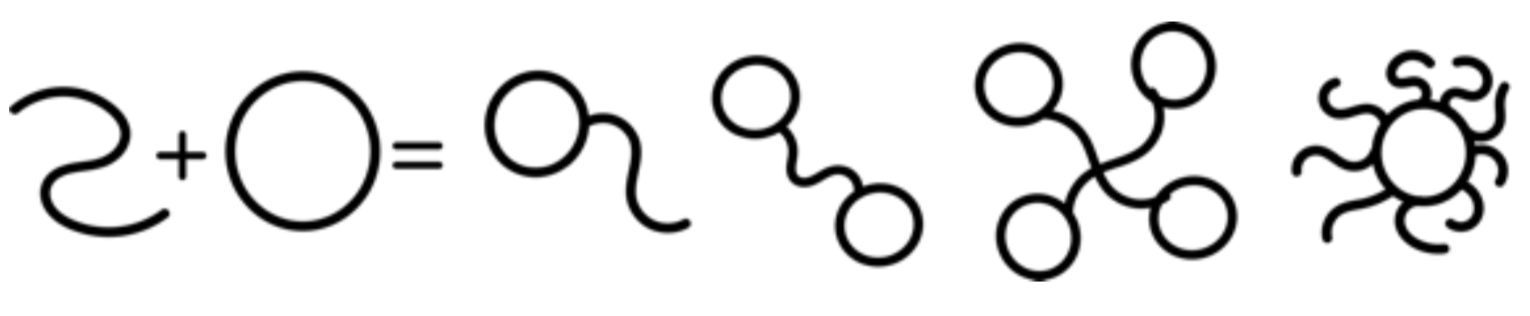 The student will be offered with 2 possible projects: (i) develop and perform simulations aimed at understanding the behaviour of non-equilibrium living polymers, i.e. undergoing architectural and/or topological operations on experimental timescales (see this paper) or (ii) perform simulations on polymers with “chimeric” topology and/or supercoiled DNA. An example of chimeric polymers can be seen in the figure to the right or this paper. The student will learn and perform Molecular Dynamics simulations of coarse-grained polymer models for DNA. S/he will develop new algorithms, run large-scale simulations on supercomputers and analyse large datasets. The grand goal of this project is to understand the relationship between polymer topology and the rheology of the complex fluid. This will unlock the possibility to (i) change the viscoelasticity of complex fluids by suitable topological operations or (ii) design fluids with desired rheology by choosing the architecture/topology of the polymers.
The student will be offered with 2 possible projects: (i) develop and perform simulations aimed at understanding the behaviour of non-equilibrium living polymers, i.e. undergoing architectural and/or topological operations on experimental timescales (see this paper) or (ii) perform simulations on polymers with “chimeric” topology and/or supercoiled DNA. An example of chimeric polymers can be seen in the figure to the right or this paper. The student will learn and perform Molecular Dynamics simulations of coarse-grained polymer models for DNA. S/he will develop new algorithms, run large-scale simulations on supercomputers and analyse large datasets. The grand goal of this project is to understand the relationship between polymer topology and the rheology of the complex fluid. This will unlock the possibility to (i) change the viscoelasticity of complex fluids by suitable topological operations or (ii) design fluids with desired rheology by choosing the architecture/topology of the polymers.
Projects are flexible and students are encouraged to discuss them with Davide in order to find the perfect fit based on the student background.
The student will work closely with other PhDs or PDRAs involved in similar projects (either computational or experiments).
Willingness to experience both computational and experimental aspects of the project is highly desired.
Project supervisor
- Dr. Davide Michieletto (School of Physics & Astronomy, University of Edinburgh)
The project supervisor welcomes informal enquiries about this project.
Find out more about this research area
The links below summarise our research in the area(s) relevant to this project:
- Find out more about Physics of Living Matter.
- Find out more about Soft Matter Physics.
- Find out more about Computational Materials Physics.
- Find out more about the Institute for Condensed Matter and Complex Systems.
What next?
- Find out how to apply for our PhD degrees.
- Find out about fees and funding and studentship opportunities.
- View and complete the application form (on the main University website).
- Find out how to contact us for more information.
More PhD projects
- Browse other Physics of Living Matter projects.
- Browse other Soft Matter Physics projects.
- Browse other Computational Materials Physics projects.
- Browse other Institute for Condensed Matter and Complex Systems projects.
- Browse all PhD research opportunities in the School of Physics & Astronomy.
- Browse PhD research opportunities elsewhere in the University of Edinburgh.

